When cells move through the cell cycle, it is often difficult to accurately assess the proportion of cells in S-phase using just a single parameter. This especially true since DNA content analyses are performed with modelling software that rely one of two algorithms (Dean Jett Fox or Watson Pragmatic) to calculate the proportion of cells in each cell cycle stage. These algorithms are imperfect in that there tends to be some error in determining the true percentage of cells in each phase due to overlap in the G1 – S and S – G2M phase transitions depending on how well the analytes fit the models.
Bromodeoxyuridine, a DNA base analogue like 3H-thymidine, can be incorporated into newly synthesized DNA when added to cell cultures. When cells are probed with a fluorescent anti-BrdU antibody, a more accurate representation of the cells in each of the cell cycle phases can be obtained and compared over a course of treatments. Changes in number of the cells in S-phase could indicate accelerated/decelerated cell division when compared to control. In the figure below, excerpted from J. Virol. 2014;88:4480-4492, a clear difference in the percentage of cells found in S phase is observed between the two methods, where the BrdU method yields a greater value in both the FLAG –ve and FLAG +ve sorting conditions.
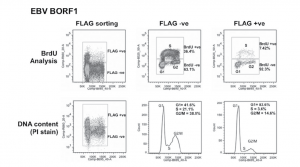
Figure 2: Cell cycle effects of EBV proteins in CNE2Z cells. CNE2Z cells were transfected with plasmids expressing FLAG-tagged BORF1 and 24 h later, were either pulse labelled with BrdU (top) or harvested directly and stained with propidium iodide (PI). Cells were divided into FLAG-positive and FLAG-negative cells by FACS and then analyzed for BrdU (S phase) and PI (DNA content) in the top panels or PI alone (bottom). Patrick Paladino et al. J. Virol. 2014;88:4480-4492
A more up to date version of this assay is the EdU Click-iT protocol developed by Molecular Probes. The harsh denaturing conditions required to expose the BrdU binding sites to antibody make it difficult to combine this assay with other probes. The EdU assay is less harsh and much faster and is a good alternative for BrdU.




