In all flow cytometric analyses, it is highly important to capture biologically significant data relative to the question being asked. As such, analysing cells that are healthy/highly viable is key to good data acquisition.
In many cases, freshly isolated cells, cells grown in culture or those treated with harsh reagents can undergo spontaneous cell death releasing their contents into the surrounding media. This material can have an agglutinating effect causing cells and antibodies to aggregate into non-specifically labelled clusters. This can be particularly confounding since not only does this make analysis of viable cells difficult, but the number of single cells for analysis in the suspension is reduced. Moreover, as cells begin to die, they become porous and expand prior to the complete loss of structural integrity and fragmentation into small vesicles or apoptotic bodies. This increased porosity also allows antibodies to cross normally impermeable cell membranes into the cytosol where they can interact with intracellular proteins via off target binding interactions. This can be problematic, especially when acquiring data for assays performed under live conditions, as dying cells may then seem to positively express the antigens of interest, however, the signals obtained may be misleading.
When acquiring data, viability can be roughly estimated from a simple plot of forward vs. side scatter. However, dead cell discrimination is not fully realized by this method, where porous cells, in early stages of apoptosis (with potential non-specific labels), can be misidentified as healthy since there is little difference in scattering patterns. It is therefore important to include dead cell discrimination dyes as part of the experimental design to minimize the inclusion of false positives when analysing the data.
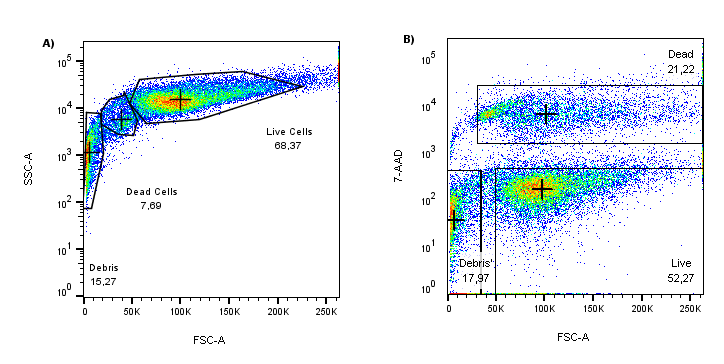
Figure 1: (A) Scatter profile of cultured HEK293T cells (FSC vs SSC). (B) When the same FSC data is plotted against 7-AAD (y-axis), unhealthy cells are nicely separated from the 7-AAD low/negative with a highly fluorescent 7-AAD population.
When comparing the FSC axes on the plots above, the 7-AAD stain reveals many cells in the process of dying that might have been included in the analysis if relying solely on scattering profiles. In this example, if scatter gating alone was used for dead cell discrimination the number of “healthy” cells analyzed would have been overestimated by greater than 10% i.e.: 7.69% in (A) vs 21.22% in (B), therefore, when planning flow cytometry experiments it is highly important to include a dead cell exclusion dye as part of the fluorophore panel.
There are many dead cell exclusions markers available across a wide range of excitation and emission wavelengths allowing for a high degree of flexibility in panel design. Traditional DNA intercalating stains such as Propidium Iodide, 7-Aminoactinomycin-D, or DAPI are impermeant to cells with intact membranes and may only be used in experiments with unfixed cells whereas, amine reactive dyes such as the Live/Dead® Fixable series (ThermoFisher) or the Zombie™ dyes (Biolegend) can be used for both fixed and unfixed samples.
For further details regarding dead cell discrimination probes and their uses in either live or fixed analysis conditions please visit our Viability Stains page here.
ThermoFishser: Checking Vital Signs: Don’t Let Dead Cells Mislead You
Biolegend: Live Cell/Dead Cell Discrimination
R&D Systems: Flow Cytometry Protocol for Analysis of Cell Viability using Propidium Iodide
Expert Cytometry: 3 Reagents for Identifying Live, Dead, And Apoptotic Cells by Flow Cytometry
BitesizeBio.com: Viability Dyes for Flow Cytometry: It’s Not Just a Matter of Life and Death
DNA Content & Ploidy Analysis
MEASURING DNA CONTENT
In normal cells, errors in DNA coding trigger cell cycle arrest in the G1/S or G2M phases and activate cell cycle associated checkpoint pathways that mediate stage dependent DNA repair. Once the damage has been repaired, arrested cells may either remain permanently arrested, re-enter the cell cycle or undergo apoptosis (if the damage is too severe). In cancer, the uncontrolled growth characteristic of the disease is associated with decreased time spent at DNA repair checkpoints, their complete bypass, poor or incomplete DNA repair and accelerated proliferation in the more aggressive forms. The search for improved cancer therapies often focuses on DNA repair checkpoint restoration, which can be studied by monitoring cell cycle progression and timing.
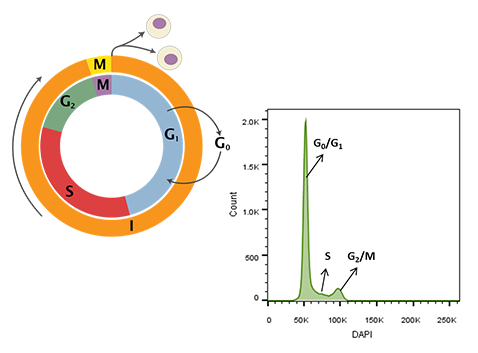
Aside from immunophenotyping, DNA content measurement is one of the most routinely performed flow cytometry-based assays. These experiments require the use of DNA intercalating dyes, such as DAPI, Propidium Iodide, or Dye Cycle Violet, that when nested between DNA bases yield signals directly proportional to the amount of DNA within the nucleus [1]. Since DNA content changes predictably with the stage of the cell cycle, the stoichiometric nature of the dye interactions with nucleic acids makes it possible to determine the cell cycle profile of cultured cells (Figure 1). For example, cells that are rapidly dividing have greater S-phase content in contrast to cells arrested in the G0/G1 or G2/M replication checkpoints [2].
Figure 1. Cell cycle schematic (left) and DAPI-intensity histogram (right) obtained from analysis of DAPI stained single cells. DNA content doubles from G0/G1 to G2/M – the first peak in the histogram at ~50K median fluorescence intensity (MFI) is representative of G0/G1 whereas the second peak at ~ 100K MFI is representative of G2/M. Cells in S-phase have variable DNA content between that of cells in G0/G1 and G2/M during active DNA synthesis. Cell cycle schematic adapted from Richard Wheeler (Zephyris) 2006.
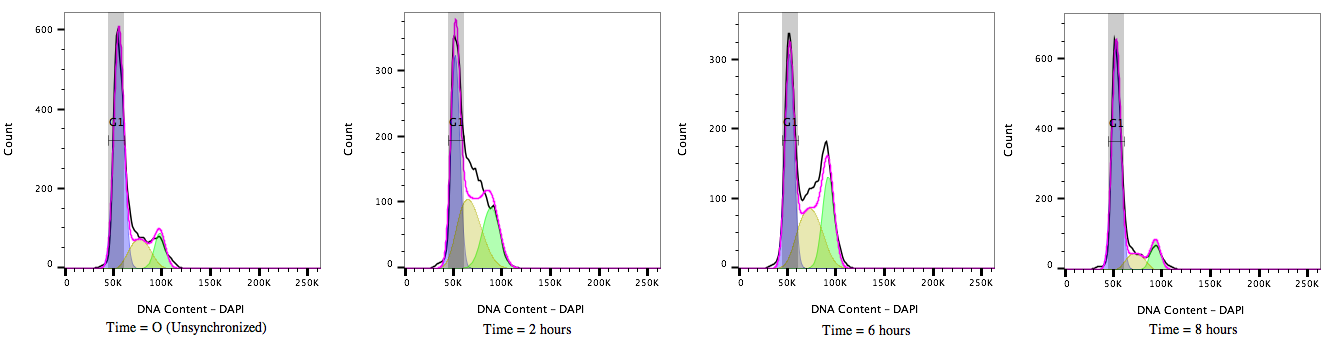
Figure 2: Cell cycle Progression of HEK 293T cells treated with a cell cycle modulating agent cells and stained with DAPI (4’, 6’-Diamidino-2-Phenylindole). Over time, from T = 0 to T= 8 hours, the cultures move through the cell cycle transitioning out of G1/0 (A) through S-phase (B) to G2M accumulation(C) and then cell division before resting again in the G1/0 phase (D).
Cancer cells and cell lines commonly have an irregular number of chromosomes (aneuploidy) caused by replication errors and the uneven distribution of genetic material during mitosis; in tumours, a greater degree of aneuploidy strongly correlates with aggressive hard to treat cancers [3, 4]. Ploidy determinations are important in characterizing tumours in the clinical setting, but are also useful in basic research when studying cancer cell drug responses in vitro.
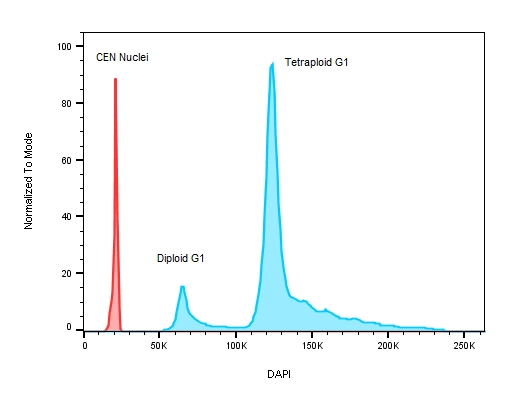
When measuring ploidy, DNA content is described as a DNA Index (DI) or the ratio of the DNA content of the experimental sample to the DNA content of a corresponding normal control (diploid) population [1]. To properly calculate the DI, an internal DNA standard must be added to each sample to ensure accurate instrument setup and DNA measurements – chicken (CEN) and trout erythrocyte nuclei (TEN) are used for this purpose since their DNA content is known to be less than that of human cells. In fact, CENs have roughly one third the amount of genomic DNA per cell compared to humans [5] making them good internal standards as their DNA profiles will not overlap with those of the tested samples, see Figure 3.
Figure 3: A plot of DNA content on a stem cell line with known replication defects causing the cells to have an abnormal DNA profile. The red peak on the histogram represents the internal control – chicken erythrocyte nuclei with lower roughly 3 x less DNA when compared to human ESC test samples where the ratio between CEN to the diploid G1 peak is 3.044 and 5.767 for the tetraploid G1.
Another useful control in the assessment of ploidy is non-proliferating human lymphocytes. In this scenario a patient’s own lymphocytes may be acquired alongside their tumour cells to measure any differences between the populations [6].
For accurate measurements, it is best to acquire data on isolated nuclei. Mitochondria multiply in response the increased energy demands of larger cells and have their own DNA and replication apparatus. As the mitochondrial burden in the cell increases this extra-nuclear DNA can increase background DNA fluorescence making ploidy measurements difficult [1].
[1] Darzynkiewicz, Z., Halicka, H. D., & Zhao, H. (2010). Analysis of Cellular DNA Content by Flow and Laser Scanning Cytometry. Advances in Experimental Medicine and Biology, 676, 137–147.
[2] Darzynkiewicz, Z., Traganos, F., Zhao, H., Halicka, H. D., & Li, J. (2011). Cytometry of DNA Replication and RNA Synthesis: Historical Perspective and Recent Advances Based on “Click Chemistry.” Cytometry. Part A : The Journal of the International Society for Analytical Cytology, 79(5), 328–337. http://doi.org/10.1002/cyto.a.21048
[3] Yildirim‐Assaf S, Coumbos A, Hopfenmüller W, Foss H, Stein H, Kühn W. The prognostic significance of determining DNA content in breast cancer by DNA image cytometry: the role of high grade aneuploidy in node negative breast cancer. Journal of Clinical Pathology. 2007;60(6):649-655. doi:10.1136/jcp.2005.035550.
[4] Rancés Blanco, Charles E. Rengifo, Mercedes Cedeño, Milagros Frómeta, and Enrique Rengifo, “Flow Cytometric Measurement of Aneuploid DNA Content Correlates with High S-Phase Fraction and Poor Prognosis in Patients with Non-Small-Cell Lung Cancer,” ISRN Biomarkers, vol. 2013, Article ID 354123, 8 pages, 2013. doi:10.1155/2013/354123
[5] Iversen, O. E. and Laerum, O. D. (1987), Trout and salmon erythrocytes and human leukocytes as internal standards for ploidy control in flow cytometry. Cytometry, 8: 190–196. doi:10.1002/cyto.990080212
[6] Benn, D. E. and Robinson, B. G. (1996), Peripheral blood mononuclear cells are a reliable internal standard for the flow cytometric DNA analysis of frozen malignant breast biopsies. Cytometry, 26: 161–165. doi:10.1002/(SICI)1097-0320(19960615)26:2<161::AID-CYTO10>3.0.CO;2-L
Learn More

Work around very talented people.
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua.
7226-1 King’s College Circle
Medical Sciences Building University of Toronto
416-976-6524